Systems Biology - Multiscale Biology
Systems Biology
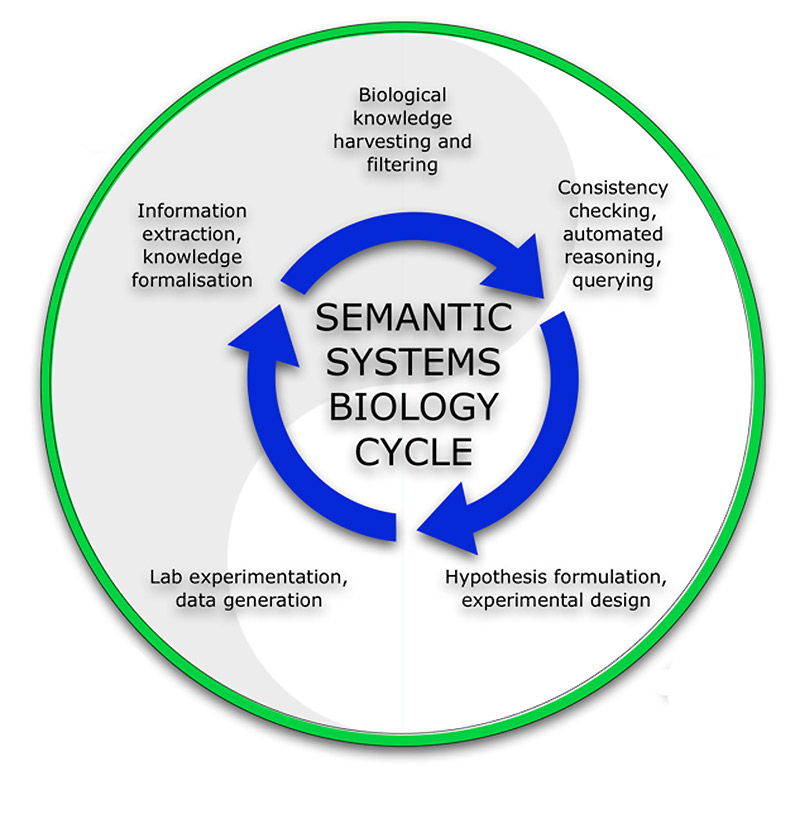
Semantic web technologies play an increasingly important role in describing and managing biological knowledge.
Whereas Systems Biology aims to provide an understanding of biological systems through the (mathematical) modelling of biological knowledge, Semantic Systems Biology (SSB) is designed to provide help in knowledge integration and network building, computational reasoning, and automated hypothesis generation and assessment. The Kuiper group collaborates with several wet-lab science groups to demonstrate the potential of SSB for the understanding of biological processes, mainly through the use of network- and pathway based analysis approaches.
The group has developed the BioGateway semantic knowledge base, which uses ontologies and semantic web technologies for the integration and exploration of biological knowledge. To provide easier access to the BioGateway knowledge, the BioGateway App was produced, allowing Cytoscape users to explore biological network building. In addition, the group extended its focus on the development of procedures and approaches to build high quality knowledge sources (the ‘Knowledge Commons’) for understanding gene regulation processes (TFcheckpoint, ExTRI, GREEKC and the Gene Regulation Consortium (GRECO)), and the production of regulatory, or ‘causal’, statements from which logical models can be constructed that can be instrumental to predict effects of system perturbations (MI2CAST).
More recently, the group has spearheaded the initiation of the DrugLogics initiative, in which several local NTNU PIs (Åsmund Flobak, Astrid Lægreid, Berit Johansen, Kaisa Lehti, Torunn Bruland, Atle Van Beelen Granlund, Liv Thommesen, Rune Nydal, Daniela Sueldo) collaborate with international PIs in the development and application of logical modelling approaches for the representation and in silico testing of several cancer and inflammation based diseases.
